Erythemato-Squamous Disease Classification Using Python
By Rachit Singh
Determining the type of Erythemato-Squamous Disease using Machine Learning Algorithms
INTRODUCTION:
Dermatology faces a serious challenge with the differential diagnosis of erythemato-squamous disorders. There aren't many differences between them other than the clinical characteristics of erythema and scaling. Psoriasis, seboreic dermatitis, lichen planus, pityriasis rosea, cronic dermatitis, and pityriasis rubra pilaris are diseases that fall in this category.
Typically, a biopsy is required for the diagnosis, but sadly, these diseases also share a lot of histological characteristics. Another challenge in making a differential diagnosis is that a disease may first resemble another disease while later developing its own distinctive symptoms.
OBJECTIVE:
To determine the accuracy of Decision Tree & K-Nearest Neighbour Classification algorithms to classify the type of Erythemato-Squamous Disease.
A. Dataset Information:
A download link for the dataset can be found at https://www.kaggle.com/datasets/syslogg/dermatology-dataset. There are 34 properties in this database, 33 of which have linear values, and one has a nominal value. There are 366 instances (rows) and 34 attributes (columns) in all. Each additional characteristic (clinical and histopathological) received a grade between 0 and 3. Here, 0 denotes the absence of the feature, 3 denotes the maximum quantity that might be used, and 1, 2 denote the relative middle values.
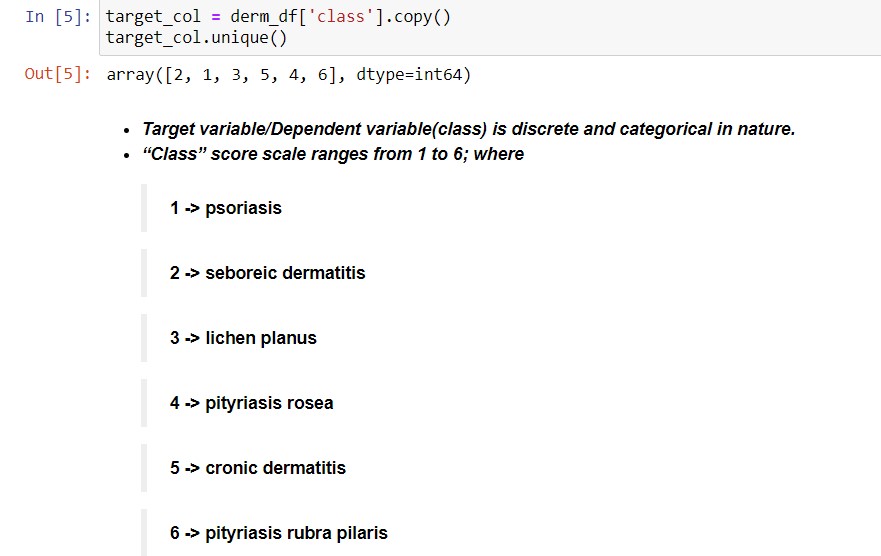
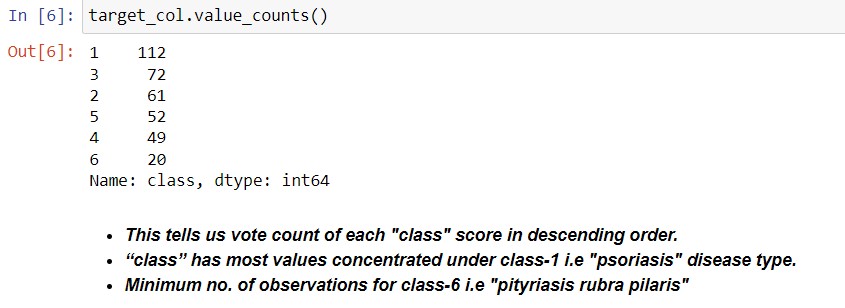
B. Class (Disease type) Distribution:
|
Class code |
Class |
Number of instances |
|
1 |
psoriasis |
112 |
|
2 |
seboreic dermatitis |
61 |
|
3 |
lichen planus |
72 |
|
4 |
pityriasis rosea |
49 |
|
5 |
cronic dermatitis |
52 |
|
6 |
pityriasis rubra pilaris |
20 |
C. Exploratory Data Analysis Insights:
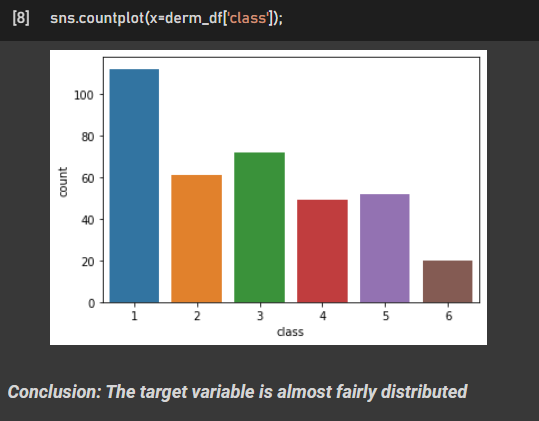
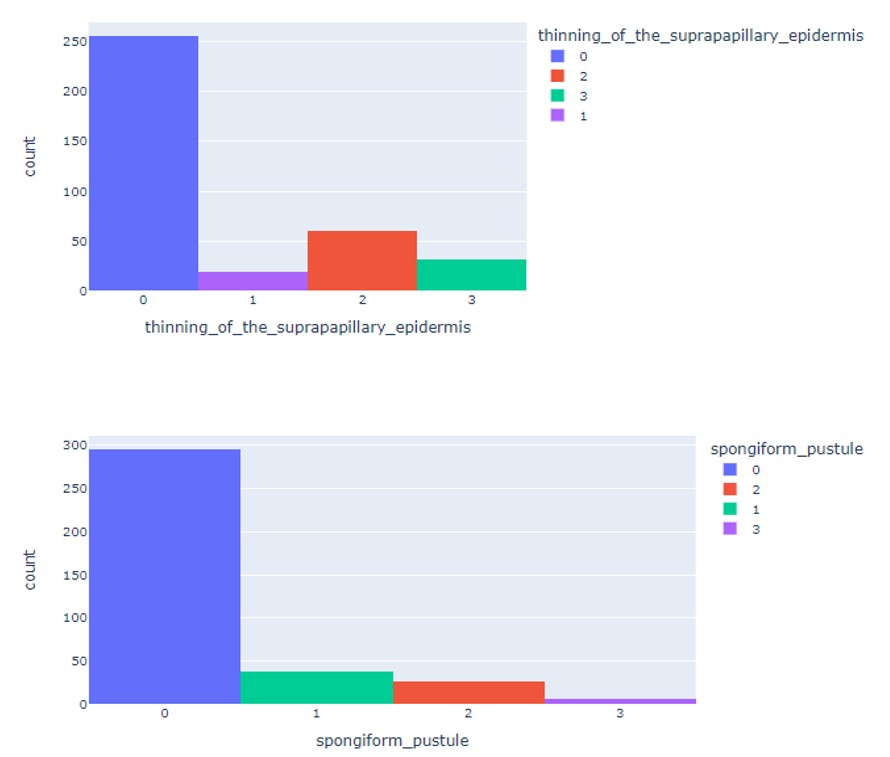
D. Machine Learning Algorithms used (Build from scratch):
Note: 80% data ( 292 records) is used for training set and 20 % (74 records) for test set.
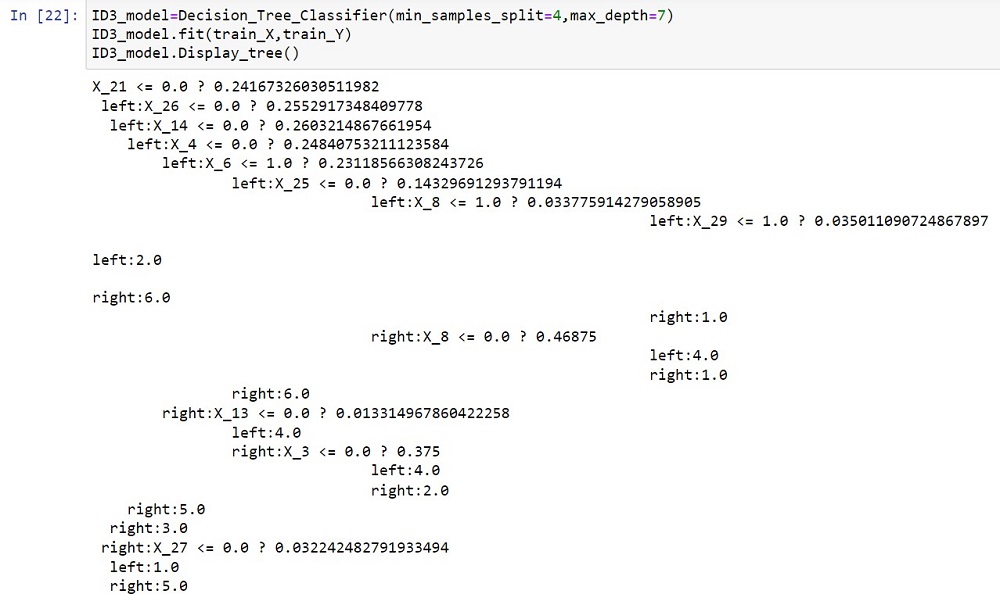
(i) Decision Tree Classifier: The algorithm creates a decision tree classifier that uses input features and labelled training data to predict outcomes. The best feature and threshold are used to recursively split the data, taking into account the minimum number of samples per split and the maximum depth of the tree. The trained model can be used to predict outcomes for new data while displaying the tree topology.
Class Node attributes:
feature_index, threshold defines the condition for the decision nodes.
left, right are edges for accessing left child and right child node respectively.
Info_gain is the information gain by the split denoted by a particular decision node.
Information gain tells us how important a given attribute of the feature vectors is.
Entropy [E]=−∑pi(log2pi)
Gini=1−∑ni=1(pi)2
Information Gain = Entropy(parent) – [Avg. Entropy(children)]
Information Gain = Gini (Parent) –∑wi Gini (childi)
Class Decision_Tree_Classifier(): It represents the decision tree model.
Constructor Attributes:
- root marks the start of Decision Tree, from which traversal takes place.
- min_sample_split, max_depth are the stopping conditions
- for e.g., if in a particular node, the no. of samples become less than the min_sample_split value, we won’t split that node further i.e., we will treat that node as leaf node.
- Also, if depth of tree reaches max_depth, then we don’t split the node further.
- The default value is set to none. This will often result in over-fitted decision trees. The depth parameter is one of the ways in which we can regularize the tree, or limit the way it grows to prevent over-fitting.
- min_samples_splitspecifies the minimum number of samples required to split an internal node,
- For instance, if min_samples_split = 5, and there are 7 samples at an internal node, then the split is allowed.
Create_tree () is a recursive function that builds tree.
- Dataframe is divided into input and target columns as X, Y variables.
- Split until stopping conditions are met.
- Then it extracts the number of samples(rows) and number of features(columns).
- Find the best split.
- It checks if Information gain corresponding to this split >0, because if info_gain=0, it means we are splitting a node which is pure (node consists of only 1 type of class).
- We create left & right subtree respectively through recursive calls. At every call we have to increase the current depth by 1.
- After creation the function should return a node which is the decision node. As it is a decision node, we have to pass feature index, threshold, left & right subtree connecters and info gain
- Now we have to compute leaf node using calculate_leaf_value ()
- At last, it should return leaf node.
Get_best_split (): It finds the best feature and threshold for splitting the data depending on the information gain. It iterates through each feature and threshold combination to assess the split's information gain.
- Create a dictionary to store best split and initialise max_info_gain as -infinity, as we have to maximise its value.
- Loop over all the possible features, inside this loop we have to traverse through all possible threshold values.
The information_gain() method computes the split's information gain. It compares the impurity (entropy or Gini index) before and after the divide.
The entropy() and gini_index() methods compute entropy and Gini index, which are impurity measurements in a set of labels.
The Compute_leaf_value() method determines a leaf node's anticipated value or class label. It selects the most common class label from the associated samples of the leaf node.
Display_tree() prints the decision tree structure, including the splitting circumstances and leaf node values.
The fit() method uses the specified characteristics X and labels Y to train the decision tree. It prepares the dataset by concatenating the features and labels and then calling Create_tree() to build the tree.
By traversing the decision tree and returning the leaf node values, the predict method guesses the labels for a given collection of features X.
The make_prediction() method walks the decision tree recursively until it reaches a leaf node, which produces the final prediction.
(ii) K-Nearest Neighbours Classifier: Predictions are made by locating the K nearest neighbours based on Euclidean distance and the model is trained by storing the training data. The majority vote among the neighbours chooses the projected label.
The KNN model is represented by the class K_Nearest_Neighbors_Classifier(), which is defined.
The value of K( taken as 4 in for this case), which establishes the number of nearest neighbours to take into account for classification, must be provided when initialising a K_Nearest_Neighbors_Classifier object.
The KNN classifier is trained using the fit() method. As input, it stores the training features X_train and labels Y_train in the object.
Making predictions for fresh data is done using the predict() method. It receives the input of the test features X_test and outputs the expected labels. The find_neighbors() method is used to iteratively search for the K closest neighbours for each test data point. The mode (most prevalent label) among the neighbours is used to establish the anticipated label.
For a given data point x, the find_neighbors() method returns the K closest neighbours. Using the euclidean approach, it computes the Euclidean distance between x and each training data point. The distances are sorted, and the labels associated with them are returned.
The euclidean() method computes the distance between two data points x and x_train. It finds the square root of the sum of squared disparities between the feature values.
RESULTS AND CONCLUSION:
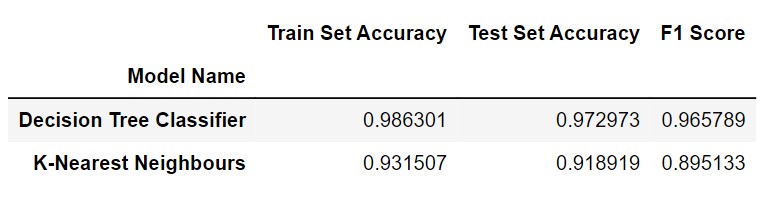
Decision Tree & K-Nearest Neighbours classification models have been applied on this dataset. The Decision Tree Classifier had highest test accuracy of 98.6301% and max F-Score of 0.965789.
Detection of skin disease is one of the major problems in the medical industry and can be healed and retrieved if properly diagnosed at an early point. Literature study demonstrates that different skin disease observation techniques are being used. However, there is still a great need to classify skin diseases at an early point. Machine learning algorithms have the potential to have an impact on early detection of skin diseases. It can assist people make real-time adjustments to their skin. If embraced well, the techniques will certainly provide appropriate assistance and a unified approach to skin problems prevention. This will assist patients and physicians cure skin diseases in a timely manner. Research and execution of limited medical information are accessible.
If more real-time data are available in the future, the detection of skin disease can be explored with recent advances in AI and the benefits of diagnosis assisted with AI.
Submitted by Rachit Singh (Rachit02)
Download packets of source code on Coders Packet
Comments