Diabetes detection using Neural Network in Python using Keras
In this project I have built a model in Python with Keras which will detect whether a person has diabetes or not using certain features. Here is a simple ANN to detect diabetes using Keras.
This project is written in Python and will require following libraries:
1.Keras
2.NumPy
3.pandas
4.scikit-learn
5.Seaborn
We will build a neural network which will be able to classify whether a person has diabetes or not.
The steps followed in the project are:
1. Exploring the dataset
2. Data visualization
3. Processing(cleaning) the dataset
4. Building the data pipeline for the model
5. Using keras to build the model and training
6. Model evaluation and tuning.
STEPS:
To begin with, let's import all the libraries.
import tensorflow.keras as keras import numpy as np import pandas as pd from sklearn.model_selection import train_test_split import os
Let's quickly import the dataset. Here it is in csv format.
ds = pd.read_csv('/content/diabetes.csv')
Then, we will go through the dataset and look at the attributes. As we can see, there are 768 samples, so the dataset is pretty small and it has 8 attributes and the 'Outcomes' column consists of the label - whether the sample has diabetes.
print(ds.info()) <class 'pandas.core.frame.DataFrame'> RangeIndex: 768 entries, 0 to 767 Data columns (total 9 columns): # Column Non-Null Count Dtype --- ------ -------------- ----- 0 Pregnancies 768 non-null int64 1 Glucose 768 non-null int64 2 BloodPressure 768 non-null int64 3 SkinThickness 768 non-null int64 4 Insulin 768 non-null int64 5 BMI 768 non-null float64 6 DiabetesPedigreeFunction 768 non-null float64 7 Age 768 non-null int64 8 Outcome 768 non-null int64 dtypes: float64(2), int64(7) memory usage: 54.1 KB None
Now, let us look at the distribution of the dataset
ds.describe() Pregnancies Glucose BloodPressure SkinThickness Insulin BMI DiabetesPedigreeFunction Age Outcome count 768.000000 768.000000 768.000000 768.000000 768.000000 768.000000 768.000000 768.000000 768.000000 mean 3.845052 120.894531 69.105469 20.536458 79.799479 31.992578 0.471876 33.240885 0.348958 std 3.369578 31.972618 19.355807 15.952218 115.244002 7.884160 0.331329 11.760232 0.476951 min 0.000000 0.000000 0.000000 0.000000 0.000000 0.000000 0.078000 21.000000 0.000000 25% 1.000000 99.000000 62.000000 0.000000 0.000000 27.300000 0.243750 24.000000 0.000000 50% 3.000000 117.000000 72.000000 23.000000 30.500000 32.000000 0.372500 29.000000 0.000000 75% 6.000000 140.250000 80.000000 32.000000 127.250000 36.600000 0.626250 41.000000 1.000000 max 17.000000 199.000000 122.000000 99.000000 846.000000 67.100000 2.420000 81.000000 1.000000
Out of the 8 features, there might be a corelation in between the features, let's check that using a heatmap. Heatmap is used to
import seaborn as sns
import matplotlib.pyplot as plt
def HeatMap(df,x=True):
correlations = ds.corr()
## Create color map ranging between two colors
cmap = sns.diverging_palette(220, 10, as_cmap=True)
fig, ax = plt.subplots(figsize=(10, 10))
fig = sns.heatmap(correlations, cmap=cmap, vmax=1.0, center=0, fmt='.2f',square=True, linewidths=.5, annot=x, cbar_kws={"shrink": .75})
fig.set_xticklabels(fig.get_xticklabels(), rotation = 90, fontsize = 10)
fig.set_yticklabels(fig.get_yticklabels(), rotation = 0, fontsize = 10)
plt.tight_layout()
plt.show()
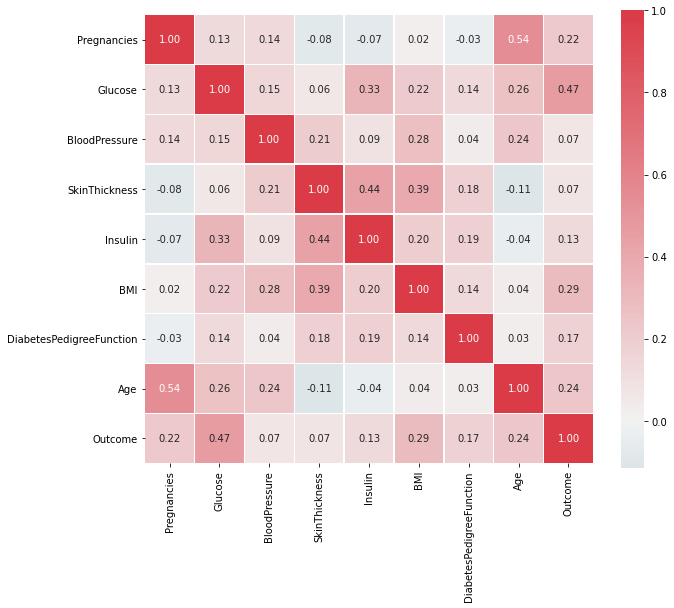
Given the dataset, there is not much of a corelation in the features.
To build the model, lets split it into train and test sets.
input = ds[columns[0:8]] label = ds[columns[-1]] x_train, x_test, y_train, y_test = train_test_split(input, label, train_size=0.8, shuffle=True)
To feed the data into the Neural Network, it should be scaled so it will be computationally easy and it will converge faster.
ip = x_train.values op = y_train.values eval_ip = x_test.values eval_op = y_test.values from sklearn import preprocessing scaler = preprocessing.StandardScaler() ip_scaled = scaler.fit_transform(ip) eval_ip_scaled = scaler.fit_transform(eval_ip)
Now let's build the model. Here, the input layer has 8 units as we have 8 features, followed by a hidden layer of 1024 units and a output layer of 1 unit. The activation layer used in the output layer is sigmoid as we will have either 0 or 1 as output.
model = keras.Sequential([
keras.layers.Dense(8, activation='relu'),
keras.layers.Dense(1024, activation='relu'),
keras.layers.Dense(1, activation='sigmoid')
])
I have used Stochastic gradient descent as optimizer with learning rate of 0.005 and Binary cross entropy as the loss function
model.compile(optimizer=keras.optimizers.SGD(0.005),metrics='accuracy', loss=keras.losses.BinaryCrossentropy())
Lets train the model
hist = model.fit(x=ip_scaled, y=op_edims,batch_size=16,epochs=1000) Epoch 1/1000 39/39 [==============================] - 0s 2ms/step - loss: 0.6881 - accuracy: 0.5586 Epoch 2/1000 39/39 [==============================] - 0s 2ms/step - loss: 0.6701 - accuracy: 0.6368 Epoch 3/1000 39/39 [==============================] - 0s 2ms/step - loss: 0.6563 - accuracy: 0.6531 ...
Epoch 1000/1000 39/39 [==============================] - 0s 2ms/step - loss: 0.3916 - accuracy: 0.8225
Here is the summary
model.summary() Model: "sequential_3" _________________________________________________________________ Layer (type) Output Shape Param # ================================================================= dense_17 (Dense) (None, 8) 72 _________________________________________________________________ dense_18 (Dense) (None, 128) 1152 _________________________________________________________________ dense_19 (Dense) (None, 1) 129 ================================================================= Total params: 1,353 Trainable params: 1,353 Non-trainable params: 0 _________________________________________________________________
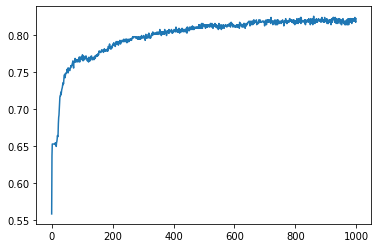
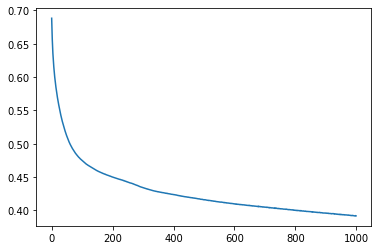
Let's plot the accuracy and the loss
Accuracy
Loss
Submitted by Saurabh Damle (saurabhdamle)
Download packets of source code on Coders Packet
Comments